DISCOVERY THROUGH GENOMICS, COMPUTATION, AND OPHTHALMOLOGY
OUR MISSION
O ur central research interest is to understand functional genetic variation in the human populations and eye diseases. We are particularly interested in characterizing how genetic variants affect the transcriptome and epigenomics, and how these molecular changes contribute to disease risk and cause phenotypic variability. We analyze these questions both by computational integration analysis of large genomic, transcriptomic and epigenomic data sets and by experimental approaches.
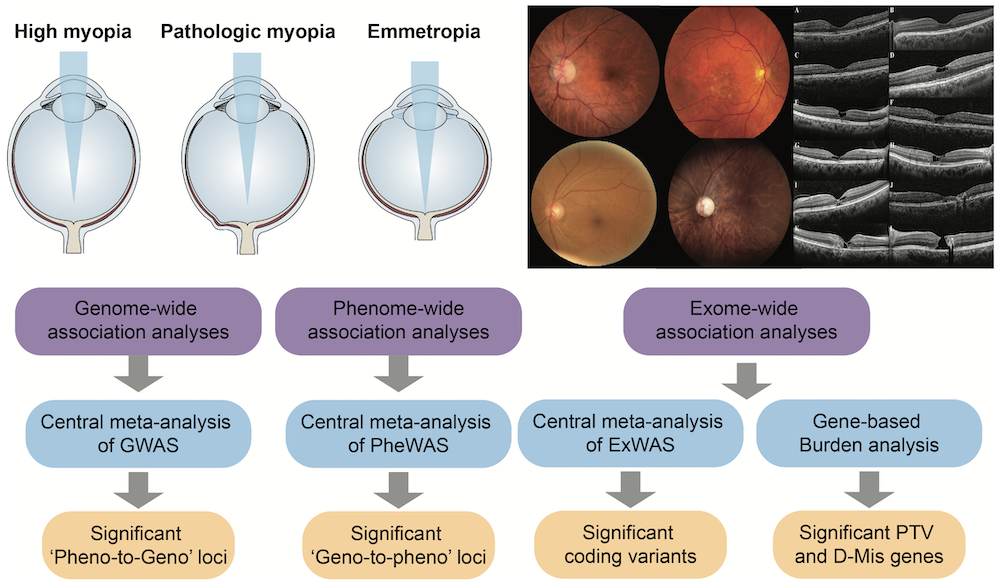
A major focus is the complex ophthalmic disorders and ocular comorbidities, where we are using color fundus photography, optical coherence tomography scanning, mouse model and retinal organoid to define clear phenotypes for mutations. We are trying to apply deep learning to create an algorithm for automated detection of medically relevant features and to develop more effective clinical diagnostic strategy and intervention methods based on these discoveries.
OUR RESEARCH
Genetic variants in human ocular phenome
D NA mutations and genetic variants have been shown to play an important role in human eye diseases and ocular traits. We’ve developed and applied genomic technologies, including whole-exome, whole-genome and RNA sequencing, to improve the diagnosis of Mendelian diseases and non-Mendelian complex diseases, with a particular focus on high myopia and retinopathy disorders (Myopia Associated Genetics and Intervention Consortium,MAGIC). Our in-house MAGIC pipeline is an a suite of bioinformatics secondary analysis tools for exploring whole-exome and whole-genome sequencing data from disease families and sporadic cases. By integrating genetic technology and expression profile, our research aims at constructing functional characterization of mutations that contribute to phenotype.
Learn More: Nature Communications 2024 Cell Reports 2023
Functional genomics of human eye disease
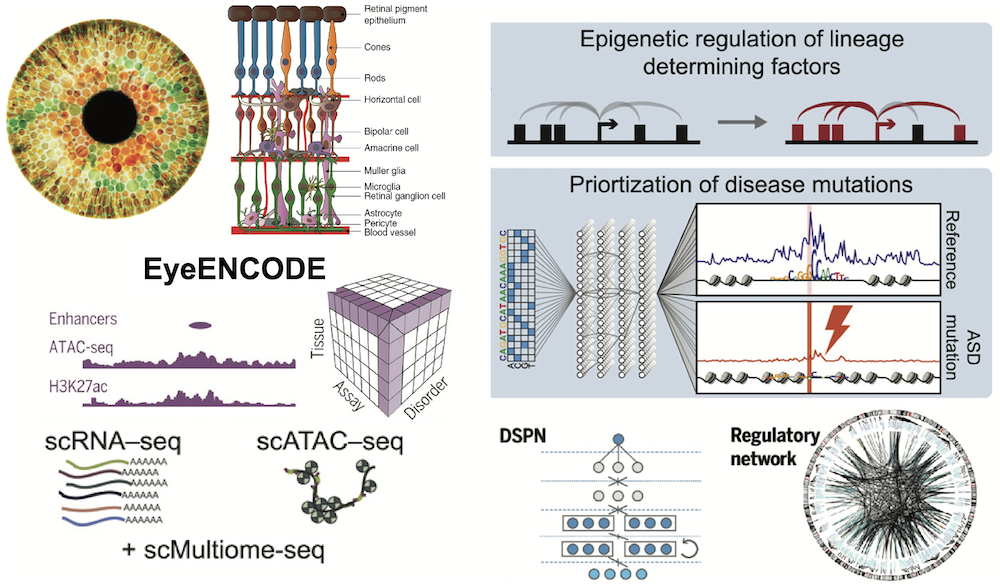
A dvances in the genetics of complex disorders reveal a highly polygenic risk architecture involving contributions of multiple common variants with small effects and rare variants with a range of effects. Because most of common genetic variation resides in noncoding regions of the genome, establishment of mechanistic links between variants and disease phenotypes is needed. We dedicated to characterize the full spectrum of genomic elements within the human eye tissue (EyeENCODE) and to elucidate their roles in development, evolution, and disease. To reach this objective, we are trying to analyze transcriptomic, epigenomic, and genomic data of adult and developing human eyes at both the tissue and single-cell levels.
Learn More: NPJ genomic medicine 2021